Thanks to Konrad Paszkiewicz from University of Exeter for this SNP-based analysis of the 3 E. coli outbreak genomes.
He used MUMmer to compare each complete E. coli genome available in NCBI to the Ec55989 chromosome, and identify single nucleotide polymorphisms (SNPs, i.e. substitution mutations, where one DNA base is swapped for another). He ignored SNPs in regions that are repeated in the Ec55989 genome, since we can’t distinguish whether these represent true substitution mutations in homologous sequences (relevant phylogenetic signal), or variation between copies in the same genome (noise). The results are available here.
He did the same comparison for the three outbreak genome assemblies, including the strain TY2482 from BGI (NCBI: AFOB00000000; Illumina HiSeq+Ion Torrent), strain LB226692 from Life Sciences (NCBI: AFOG00000000; Ion Torrent), and strain H112180280 from the UK Health Protection agency (454 mate pair). In addition he did the same comparison for the first MIRA assembly of TY2482 (Ion Torrent) and I analysed SNPs in Ty2482 using the Illumina HiSeq reads released by BGI (using bwa and samtools).
Konrad then used the annotation of Ec55989 to identify the coding change cause by each SNP – i.e. whether it is in a gene, and whether the DNA base change results in a change in the encoded protein sequence (non-synonymous SNP) or not (synonymous SNP). See resulting table here.
I used this annotation to remove SNP calls in genes annotated with the keywords ‘phage’, ‘transposase’, ‘insertion sequence’ or ‘IS’ in an effort to remove obviously horizontally transferred DNA…since this is not only subject to divergence by mutation (the phylogenetic signal we are looking for) but also to recombination. The result is just over 400,000 SNPs (nearly 10% of the Ec55989 chromosome). The alignment of those SNPs is available here.
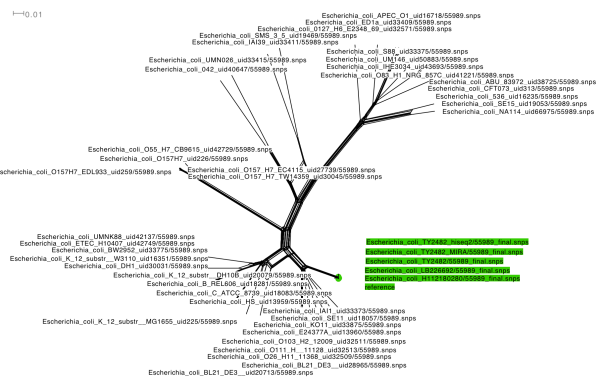
I used SplitsTree4 to draw a phylogenetic network for the alignment. The result clearly shows that the outbreak genomes (green) are very similar to Ec55989 (also green, labelled ‘reference’), and very different to other sequenced E. coli (note in particular the group of EHEC O157:H7 on the middle left, which are very distant from the outbreak strains). Since this is a network, it would reveal if there were major recombinations between this strain and the other E. coli chromosomes, which there aren’t. This confirms that the outbreak strain is truly an EAEC, with very close similarity to Ec55989 and not to classical EHEC. This is backed by the presence of an EAEC plasmid, carrying aggregative adhesion fimbrial cluster I (AAF/I; agg operon; see this post for details).
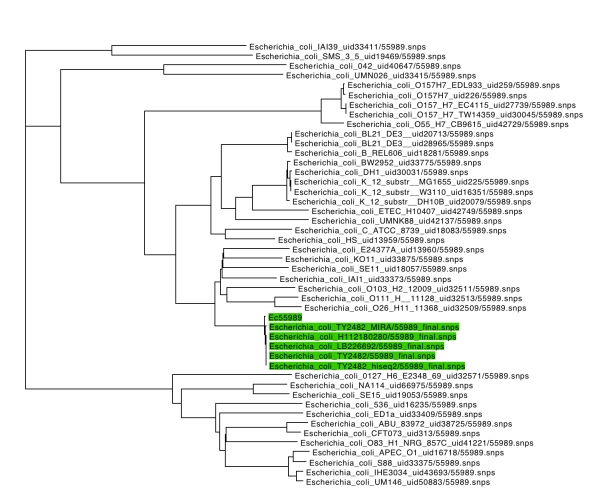
I also used SplitsTree4 to draw a phylogenetic tree of the data, using K2P distances and the BioNJ tree-drawing algorithm. This shows the same result, with all three outbreak strain’s genomes clustering very tightly with the EAEC strain Ec55989.
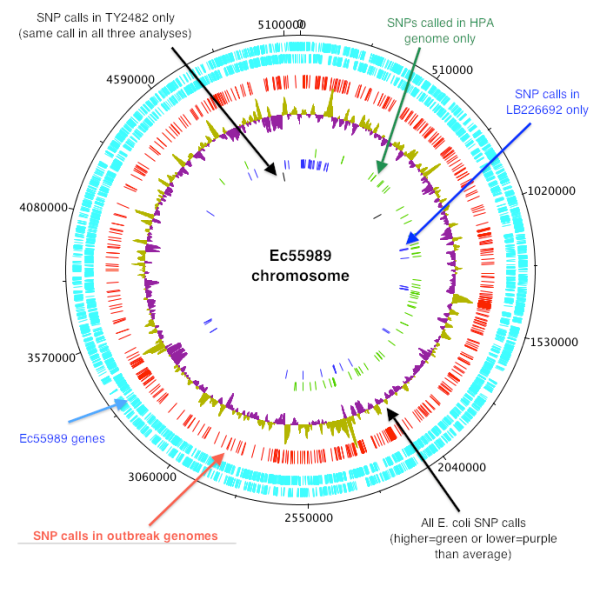
So where are the SNPs located? This figure shows the distribution of SNPs around the Ec55989 chromosome:
The outer blue rings indicate the Ec55989 genes encoded on the forward (outermost) and reverse strands. The purple/green wiggly line indicates the full set of SNPs found among all avaialable E. coli chromosomes in NCBI…this is a relative plot, so purple bits indicate where SNPs are relatively rare (lower density than the average across the genome) and green bits indicate where SNPs are particularly common (higher density than average across the genome).
The red ring indicates the location of SNPs that are shared among the three outbreak genomes compared to Ec55989, it that distinguish the outbreak from Ec55989 (~600 SNPs, or 0.12% divergence). For comparison, commonly circulating Salmonella enterica serotype Typhi, which are considered a tight-knit clonal group compared to other Salmonella and cause typhoid fever as opposed to the gastroenteritis caused by most Salmonella enterica, can differ from each other by >600 SNPs. So the outbreak strains should be considered quite closely related to the EAEC diarrhea strain Ec55989. In fact, some of these SNPs (red ring) are clustered together, suggesting that they actually represent variation in horizontally transferred sequences such as phage rather than genuine substitution mutations…so the real number of substitution mutations differentiating the outbreak strain from Ec55989 is probably more like 200-300, or 0.05% divergence.
The inner rings (green, blue, black) show the location of SNPs that are specific to just one of the three outbreak genomes. For the HPA and LB226692 strains these are VERY likely to be false SNP calls, due to the fact that they are reliant on Ion Torrent and 454 data which have trouble distinguishing between homopolymeric tracts (e.g. GGGTTT can easily be misread as GGTTTT, masquerading as a substitution of G->T). For TY2482, where we have Illumina read data to call SNPs (which doesn’t have this homopolymer issue), we find only 2 SNPs that are unique to Ty2482 compared to the other two outbreak strains (black inner ring). This is easier to believe, as we expect very small numbers of mutations to accumulate during the few weeks of evolution that separate these sequenced isolates.
Hi,
great job. Just one question for clarification. When you campare your SNP map in Splitstree4, what do you compare it to? All public e.coli genomes? How many genomes are there and how good do these represent E.coli species?
thank you
Daniel
Hi Daniel,
The E. coli genomes used for comparison in the split network are the same as those in the tree…you can see the names of the genomes in the tree figure, and also they appear in the first line of the SNP table that is linked to for download.
These represent all finished E. coli genomes currently available in NCBI. They will be biased to types of E. coli that us humans are interested in, like human and animal pathogens, and not representative of the whole genus (which as massively diverse!).
Hi Kat,
Could you please clarify in the above DNAPlotter graphic whether the red ring refers to only filtered-out-for-horizontal coding SNPs found consistently in all three assemblies or something else? It might also be helpful to provide the number of such validated cSNPs relative to the total number of proteome amino acid positions.
Now some of the established cSNPs will be neutral meandering within the amino acid reduced alphabet defined by the comparative genomics of the respective protein orthology class, others might be adaptive and so clues to selective pressure within livestock reservoir, while the rest might represent initial stages of pseudogenization and so clues as to what is not under selection. Have you broken out the cSNPs yet with respect to PolyPhen and the like?
Thanks for all you are doing here!
Tom
Hi Thomas,
Yes the red ring shows SNPs with simple filtering to remove horizontal elements (keywords ‘phage’, ‘transposase’, ‘IS’, ‘insertion sequence’). The SNPs are not just in protein coding sequences, so to answer your question you’d need to count how many were in genes, then how many amino acids there are in Ec55989 excluding those annotated with the keywords I described, and divide one by the other. To make things simpler, I just reported the total divergence:
Re analysig of coding SNPs, see above reply to PatrikD. Basically I wouldn’t bother at this point, because many will be errors. We need to clean up the SNP calls further before it’s worth doing that. The purpose of the post was really to show that, at the SNP level (as well as the gene content level which has been presented earlier) the outbreak genome is very close to Ec55989 and not to EHEC or any other E. coli in the database.
Some further comments to your most excellent post:
It appears you have laid out all the ingredients above to cook up an idealized reference genome (pooled assembly) — is there a link to that yet? As you note, the isolates are too close in time and source to have substantive information on strain evolution, so what we are reconciling here — in time-wasting triplicate — is predominantly homopolymer error. If mate pairs or contig tiling reads are available from one of the assemblies and help with another, by all means let us use them to build finished scaffolds and focus all the annotation effort on that. Surely the pooled coverage is excessive at this point.
I would draw a parallel here to the human reference genome which is also drawn on pooled dna from recently diverged strains (the donors). Rather than toss it to start over with a single individual, given the average human carries a few hundred serious mutations, there have been calls to idealize the current reference genome (replace internal stop codons with the amino acid found in healthy individuals, perhaps even replace the <1% coding SNPs as well since correlations are lost upon pooling.)
One straightforward application (for you!) of this pooled assembly to the E. coli pangenomic picture would be replacing O157:H7 with O104:H4 at the center of the 61 whole genome alignment at the first link below, which would give a very helpful revision of their Fig5 (second link, best viewed at full resolution). However you note in an earlier post that syntenic relations used there are shuffled too much for gene order to be that helpful. So it might make more sense to sort first by presence/absence relative to O104:H4 and sort the remainder by relative percent identity.
This would have the effect of reordering DNAPlotter output so that the display was more informative for evolutionary distance at a given display resolution, in the process creating usefully ordered gene fasta files. (As the fasta header, when properly constructed, is a little flatfile database in itself, this order and physical order just amount to two of the dozen or so fields.)
http://www.ncbi.nlm.nih.gov/pmc/articles/pmid/20623278
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2974192/figure/Fig5/
A few days back you promised to add a bit more legend to a DNAPlotter graphic, the one that showed separate circles for main chr, plasmids and phage. The issue there was the inner ring which seems to show GC skew, defined as (G-C)/(G+C) but subject to an unfortunate typo in the DNAPlotter manual (GC)/(G+C). Additional confusion with the strand GC composition ring might occur as you did not show all 8 rings that DNAPlotter offers. The significance here being the sign switch in DNA skew is sometimes taken as indication of plasmid replication origin, which becomes interesting relative to a later post proposing one of the plasmids was integrated into the main chr — how far back might be timed by loss of skew. http://www.sanger.ac.uk/resources/software/dnaplotter/
Hi Thomas,
I agree it would be useful to use all the available reads to create a single meta-assembly of the outbreak strain. Unfortunately HPA have removed the 454 mate pair reads which they originally posted, so this can’t be done at present…except by the folks at HPA!
Why don’t you have a go at creating the figure you suggest?
I would use BRIG.
Damn – too bad about the HPA reads! There’s still the 8 Ion Torrent runs from Life Tech, though, right? That would more than double the BGI Ion Torrent data.
Then again, I assume BGI are continuing to work on their own assembly as well. I’m sure they would prefer to close the genome entirely based on their own in-house generated data, rather than having to credit Life Tech for part of it.
Nice work as usual. You mention ~600SNPs between 55989 and the three outbreak strains, but I only counted 389? I’m looking at file http://bio-ruby.ex.ac.uk/ecoli_outbreak/updated_snp_phylogeny2/55989_snpcomparison_with_genes.txt, columns “reference” versus “Escherichia_coli_H112180280/55989_final.snps”, “Escherichia_coli_LB226692/55989_final.snps”, and “Escherichia_coli_TY2482_hiseq2/55989_final.snps”, right?
There’s obviously not enough divergence to formally calculate Ka/Ks ratios, but it may still be interesting to point out some genes with multiple SNPs:
2 or 3 non-synonymous SNPS:
EC55989_3283 conserved hypothetical protein
EC55989_3284 conserved hypothetical protein; putative exported protein
EC55989_0326 putative deaminase/amidohydrolase with metallo-dependent hydrolase domain
EC55989_1414 Putative tail fiber assembly protein
EC55989_2110 Tail protein X (GpX)
EC55989_2113 hypothetical protein
EC55989_2116 putative phage tail protein S
EC55989_4152 putative PTS system alpha-glucoside-specific EIICB component
EC55989_2207 High-molecular-weight nonribosomal peptide/polyketide synthetase 2 (HMWP2)
EC55989_4816 valyl-tRNA synthetase
2-4 synonymous SNPs:
EC55989_1076 Host specificity protein J
EC55989_3293 conserved hypothetical protein
EC55989_2128 Major tail tube protein FII
EC55989_2364 conserved hypothetical protein; putative outer membrane protein
EC55989_2130 Gene late control D protein
EC55989_4669 putative Iha adhesin ( receptor) (1 non-synonymous, 4 synonymous)
high number of SNPs:
EC55989_1411 putative tail fiber protein (4 non-synonymous, 3 synonymous)
EC55989_2595 hypothetical protein; putative head-binding domain (4 non-syn, 5 syn)
The tail fiber / tail tube / head binding proteins are all phage, so subject to horizontal transfer.
The high ration of synonymous to non-synonymous SNPS in Iha adhesin, a virulence factor in urinary tract infection, seems interesting though.
Hi Patrik, there are actually 3 columns for the TY2482 strain: “Escherichia_coli_TY2482/55989_final.snps”, “Escherichia_coli_TY2482_MIRA/55989_final.snps”, “Escherichia_coli_TY2482_hiseq2/55989_final.snps”, which were run using the various assemblies of TY2482.
If you do this:
head -n 1 55989_snpcomparison_with_genes.txt | awk ‘{print $3,$19,$28,$43,$44,$45}’
you’ll see that these are the relevant columns to compare
“reference” “Escherichia_coli_H112180280/55989_final.snps” “Escherichia_coli_LB226692/55989_final.snps” “Escherichia_coli_TY2482/55989_final.snps” “Escherichia_coli_TY2482_MIRA/55989_final.snps” “Escherichia_coli_TY2482_hiseq2/55989_final.snps”
If you do this you can count the number of SNP calls where one of these columns differs from the reference DNA base:
awk ‘$3 != $19 || $3!=$28 || $3!=$43 || $3!=$44 || $3!=$45’ 55989_snpcomparison_with_genes.txt | wc
I get 775 lines, i.e. 774 SNPs (first line is header)
If you do this on a file where all SNPs genes annotated with ‘phage’, ‘transposase’, ‘IS’ or ‘insertion sequence’ are removed, you get 650 SNPs.
But, this is an overestimate as it still contains SNPs in prophage genes which don’t include the keyword ‘phage’ in the annotation, like the ones you pointed out (e.g. EC55989_1411 putative tail fiber protein, EC55989_2595 hypothetical protein; putative head-binding domain). So these should really be removed too. I will do this at some point, or you could!
Furthermore, a large proportion of these SNPs will turn out to be errors, since it is supremely difficult to get accurate SNP calls from assemblies based on Ion Torrent or 454 data due to their trouble with homopolymeric sequences. So I would caution strongly against any sophisticated analysis of the data presented here! The point of posting this was to show that the genome is very very similar to Ec55989 and very different from others…finer detailed analysis of the mutations will require careful analysis of the read data. If you want to look at the locations of SNPs, I would recommend using the data in the Ty2482 HiSeq column as this will be the most reliable of the SNP calls of the outbreak strain vs the Ec55989 reference.
Ah – I see! That wasn’t very clear from your post actually. You mentioned “SNPs that are shared among the three outbreak genomes compared to Ec55989”, so I looked for SNPs that were identical between H112180280, LB226692 and TY2482, but differed from 55989. I also used only the latest TY2482 since it seem to be quite a bit more reliable (that’s column TY2482_hiseq2, right?)
Since I’m looking for SNPs that agree between the three assemblies, I’m not too worried about remaining sequencing errors. Given how closely these strains were sampled, I’m not interested in any minor differences between them, but more in the difference between 55989 and the three outbreak strains.
I think the best way to do that at this point would be to use the SNP calls from HiSeq reads of TY2482.
I can’t post much in the comments so will have to write a new post… https://bacpathgenomics.wordpress.com/2011/06/15/point-mutations-snps-in-outbreak-strain-relative-to-ec55989/
Pingback: SNP-base phylogeny confirms similarity of E. coli outbreak to EAEC Ec55989 «
Pingback: Point mutations (SNPs) in outbreak strain relative to Ec55989 « bacpathgenomics
Hi there,
i am not an expert in enterobacteria, but i asked myself, if the stx-genes really derived from an EHEC strain. Afaik stx phages appear in a number of other species. So, judging from sequence similarity, what is the origin of the phage acquired by the outbreak strain? I
Thank you for helping out.
As you mention, the stx2 genes are carried by a phage. This is a sort of virus that has its own genome sequence, which is packaged up inside a protein capsule. When the phage particle meets a bacterial cell, it injects it with a copy of its own DNA sequence, which can be integrated into the bacterial chromosome. It is this DNA that contains the genes encoding Shiga-toxin. So, it is the movement of the phage that spreads the Shiga-toxin, and these phage are not restricted to EHEC. So no, we cannot say whether the stx genes came from a phage that was resident in EHEC or from a phage that was resident in some other E. coli.
In this post from last week, I showed a tree of phage sequences with homology to that in the German O104:H4 genome (using from NCBI blastn). The closest phage sequences are those from EHEC O157:H7, but there are differences, as discussed in this post by Nico Petty.
Pingback: How Genetic Engineering May Have Created E. Coli Outbreak « Wake-up Call
Pingback: How Genetic Engineering May Have Created E. Coli Outbreak « Only Ed