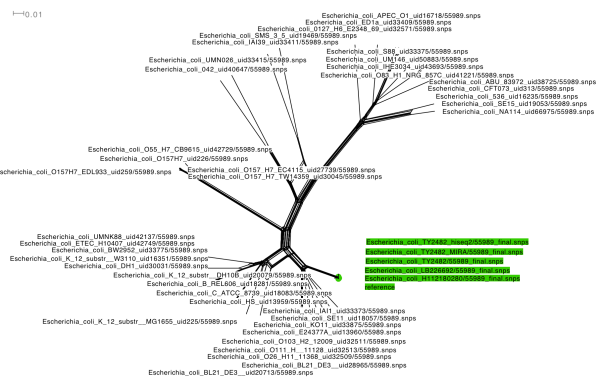
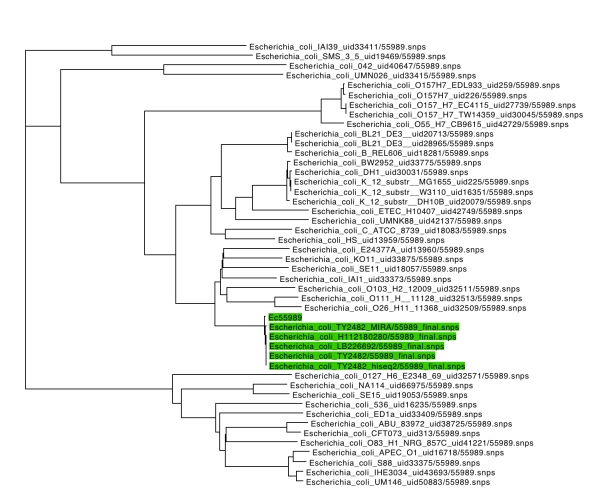
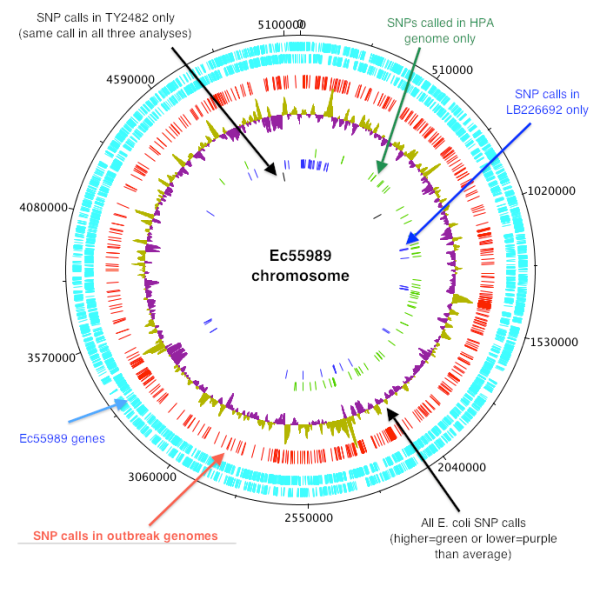
Since the Illumina (MiSeq) reads from 5 HPA genomes were recently released, I thought it would be interesting to compare these to the ‘complete’ assembly of TY2482 from BGI and look for substitution mutations and read depths for plasmids vs chromosome.
SNP analysis
I decided to try using Nesoni, the python-based analysis pipeline written by Paul Harrison & Torsten Seeman from the Victorian Bioinformatics Consortium here in Melbourne. It uses shrimp2 to map reads, and samtools and to generate and process sorted alignments and call variants. It has a nice feature where, with a single command (nesoni nway) you can generate a table showing an n-way comparison of consensus allele calls in a set of genomes, at each of the loci called as a variant in any genome.
The complete resulting output is here. It reports not only the consensus call, but the evidence behind the call, so it’s simple to see whether you believe it or not. The result is 220 calls, of which I might believe 9 (pink in figure below, and Excel file here).
I mapped MiSeq reads from 4 of the 5 HPA genomes (shrimp2 didn’t like the fastq file for sample 280, need to sort this out) and the HiSeq reads from BGI’s TY2482, to the complete reference assembly for TY2482. For 194 of the 220 variants called, the TY2482 read mapping resulted in a variant call compared to the TY2482 reference, which means that the variant is unlikely to be real. This could happen for a variety of reasons relating to the mapping & variant calling process, and I was just using the default settings in Nesoni so some tweaking might remove these. In any case, I will ignore these variants for now because I don’t believe they are real (but you can see the full table here).
This leaves 24 variant calls, where the allele detected in one or more of the 4 HPA genomes is different from the TY2482 assembly+reads from BGI, shown in the table below. This includes 9 in the chromosome (highlighted in pink), with 2 SNPs called in all 4 genomes; 1 SNP called in sample 283 only, 3 SNPs called in sample 540 and 3 SNPs called in sample 541.
In pTY1, which is the IncI plasmid bearing the beta-lactamase CTX-M-15 gene, the variants detected (yellow above) were all within the shufflon proteins…this region is able to ‘shuffle’ via inversions between homologous sequences, and these variant calls will most likely represent shuffling in the region rather than point mutation. In fact, the alignment suggests that there are multiple versions of these sequences in each DNA sample, suggesting that the shufflon was active and generating a mixed population of plasmids in the HPA data (but not the BGI strain TY2482, which had homozygous calls in this region). This might be worthy of some further investigation by someone who understands shufflons a lot better than I do!
Finally, there were a few variant calls in the tiny plasmid pTY3, clustered within its rep gene. These calls are heterozygous (see table) in all four HPA strains, suggesting that the mapping is picking up two different versions of the rep gene, which could be due to homology with other replication proteins in plasmids pTY1 and pTY2.
Plasmid copy number
The massive variation in read depth for plasmid sequences compared to the chromosome reminded me it might be interesting to try to infer the average copy number for each plasmid based on read depth. To do this, I used the depth plots output by nesoni (which gives the mean read depth per base in the reference sequence, based on read mapping). I calculated the mean read depth across each reference sequence (ie the completed BGI TY2482 assembly, chromosome + 3 plasmids) from this, and then calculated the ratio of read depths for plasmid:chromosome. Assuming each bacterial cell has ~1 copy of the chromosome (i.e. ignoring cells caught in the act of replication when there will be >1), this should give an approximation of the mean copy number of each plasmid per cell. We know some plasmids are maintained quite stably at 1 per cell, while others can exist at high copy number. This is the result:
So for the TY2482 data, it looks as though the IncI1 resistance plasmid (pTY1) and the aggregative adhesion plasmid (pTY2) are maintained at ~1 per cell, while the mini plasmid (which contains little more than a plasmid replication gene) is present at ~9 per cell. This is pretty much in line with expectation.
Interestingly, the HPA strains appear to have much higher copy numbers, around 20 per cell for pTY1 and pTY2 and hundreds of copies of pTY3. The numbers are pretty consistent across the HPA strains, but are remarkably higher than in TY2482.
I don’t have a good explanation for this apparent difference…. it could be an artefact in the sequencing (MiSeq likes plasmid DNA???) or in the mapping (not sure how this could be, especially since the mean depth plots produced by nesoni exclude regions that map to multiple locations in the reference genome).This could be examined by looking at results from different mapping programs, or analysis of reads from different platforms (Ion Torrent for TY2482 & LB226692, 454 reads for the HPA & C2L genomes).
If it is a real difference, I wonder if it could be differences in growth rate or culture conditions in the two labs. Or a mutation in the chromosome that affects the normal control of plasmid replication? Could having lots of copies of the aggregative adhesion plasmid enhance virulence or transmission of the bug?