This is a guest post from Dave Savage, a post-doc working on resistance gene analysis at the University of Melbourne. We have been trying out SPAdes as a replacement for Velvet + Velvet Optimizer which we routinely use for assembling bacterial genomes. The SPAdes paper, and the B-GAGE assembler comparison, show that SPAdes does better than most other assemblers on a small set of bacterial read sets, but we routinely assemble hundreds of genomes so needed to see whether this performance holds across large sets of reads with variable coverage etc. The results confirm SPAdes consistently performs well, and better than Velvet. This, and the fact that SPAdes gets you from fastq reads to error-corrected contigs and scaffolds in a single command with no need for tuning parameters, makes it a clear winner for our purposes.
Dave says:
SPAdes (http://bioinf.spbau.ru/spades) is a new(ish) genome assembly program that is particularly suitable for assembly of bacterial genomes. SPAdes makes a number of improvements on the de Bruijn graph algorithms used by assemblers like Velvet, IDBA and SOAPdenovo, including iteration over values of kmer sizes, and incorporation of paired kmers (k-bimers), which allows information from paired end reads to be introduced into the computation at an earlier stage.
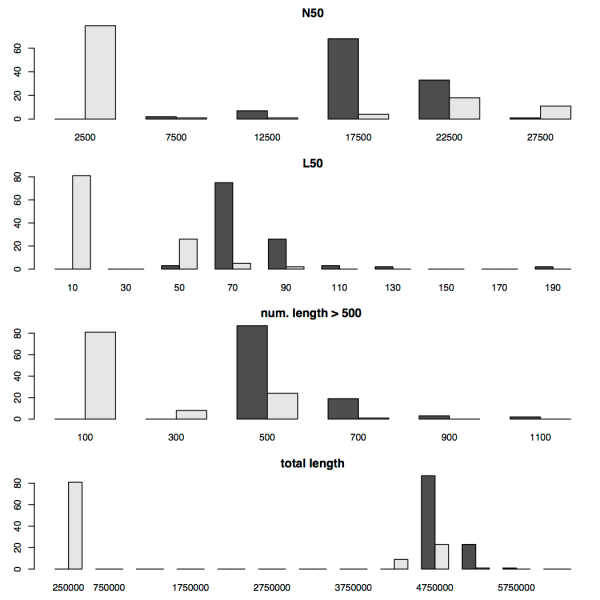
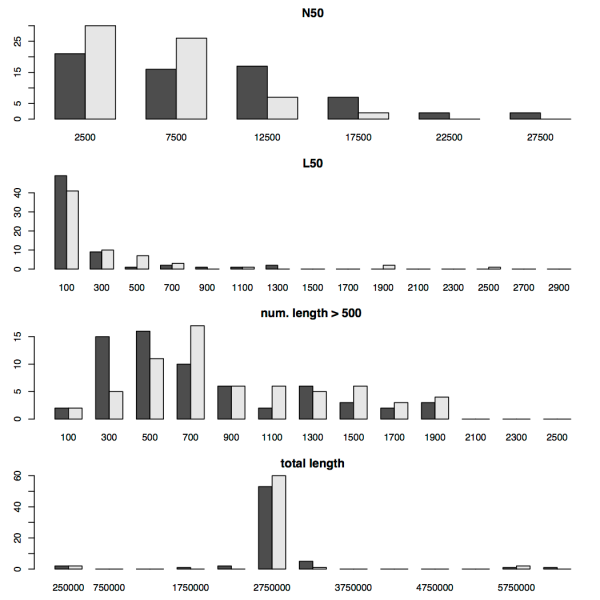
In their paper, the authors of SPAdes make comparisons with a number of popular assemblers using single-cell and multi-cell E. coli datasets. Their results show that SPAdes can in fact deliver better assemblies, with a higher N50 length and larger contigs. Since we regularly use Velvet for assembly, we wanted to perform some further comparisons between Velvet and SPAdes on the types of data sets we typically deal with. To this end we ran SPAdes and Velvet on 114 Shigella sonnei 54 bp paired end Illumina read sets (from our 2012 paper, reads available at ERP000182), and 67 unpaired 37 bp Staphylococcus aureus samples (from Harris 2010, available at ERP000070). We then calculated the N50, length, L50, the number of contigs > 500 bp, and the total contig length produced by each assembler. The figures below show the distributions of each of these values for the two data sets.
There’s a few things to notice about these plots. Firstly there’s quite a bit of difference between the S. shigella and the S. aureus assembly statistics, although this is hardly surprising given that the S. sonnei reads are paired ends, while the S. aureus are unpaired (and only 37 bp long). What’s apparent though is that SPAdes and Velvet perform similarly on the S. aureus reads, but have very different performance on the S. sonnei reads, where SPAdes clearly outperforms Velvet. We can probably attribute this to the fact that SPAdes includes an improved algorithm for handling paired end reads.
For both species, SPAdes frequently results in a higher N50 than Velvet and a larger total contig length. For the paired end S. sonnei reads SPAdes also results in a larger number of contigs greater than 500 bp in length. For the L50 metric, Velvet results in quite low scores for the S. sonnei reads, with 10 or less contigs frequently accounting for at least half of the total contig size. However, given the total contig length obtained using SPAdes, the much higher values for L50 obtained using SPAdes are not unexpected.
In our current assembly pipeline, we run Velvet Optimizer a number of times using smaller and smaller steps for k in order to hone in on an optimal value. Using SPAdes, not only is this step incorporated into the assembly program, but the information from each iteration is combined to produce the final output. Thus SPAdes is able to capture the information obtained using small, highly sensitive kmer sizes, as well as large, highly specific, kmer sizes, and at the same time simplify our assembly pipeline. We’re currently beginning a new project looking at the pan-genome of a number of bacterial species, and we anticipate that this project will involve quite a bit of assembly. Based on the results shown above, and the pipeline process for SPAdes, it looks like we’ll be using SPAdes to do the bulk of this work.
Run specifics:
- Velvet was run on the S. sonnei pared end reads using VelvetOptimser with a minimum kmer size of 29 and a maximum kmer size of 89.
- SPAdes was run on the S. sonnei pared end reads using the default settings.
- Velvet was run on the S. sonnei pared end reads using VelvetOptimser with a minimum kmer size of 15 and a maximum kmer size of 35.
- SPAdes was run on the S. sonnei pared end reads using the kmer sizes of 15, 23, 35.