I’ve been working with large, whole genome phylogenies a lot lately, and wanting to overlay metadata associated with taxa in the tree.
For example, I have a table of resistance genes and mutations detected in each bacterial strain, and a phylogenetic tree showing the relationships between strains. I want to quickly and easily plot the tree and data together, so I can see whether the resistance genes are clustered together in a single clade or lineage, or if they are cropping up in lots of unrelated strains.
There are quite a few tools out there that can do something like this, but they all seem to have their drawbacks and issues, so I ended up hacking up an R script to do what I want. Here is a quick round-up of the tools I found, and the script I came up with.
Update (28/6/12): Just spotted a new online tool, EvolView, for plotting trees and data: http://www.evolgenius.info/
iTOL
Otherwise known as interactive Tree Of Life. This is probably the one that most people know, it is a good tool and doesn’t require any programming. You just upload a newick tree file and tables of data in various formats (described here), and it can display all kinds of data (see examples on the front page). Figures can be exported in PDF and other formats, which is great for publications.
The data has to be the correct format for iTOL before uploading. For my purposes (data = gene content or mutations) I usually use the heatmap or color-strip data types. Color-strip types are good for showing categorical variables, e.g. I often want to look at the distribution of different types of gyrA mutations that confer reduced resistance to fluoroquinolone-based drugs, and I need to have each mutation in a different colour. It is also good for showing the geographical location of isolates… for this, I like to use the ‘colour branches’ option, so that rather than displaying a box next to each strain indicating its location, the branches are coloured according to the location of the leaves below them…this is a good way to highlight clades within the tree that are geographically clustered (see the example below). To convert a simple table (column 1 = strain names, which match the leaves of the tree, other columns = categorical variables) into one suitable for uploading as a colour strip to iTOL, I wrote this simple Python script.
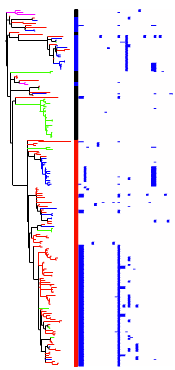
Here is an example with resistance mutations shown as a colour strip (red/blue/black), plasmid coverage shown as a heatmap, and geographical locations indicated by branch colours (achieved using the ‘colour strip’ data type). Note that this is a screenshot from my browser, as the export option was not functioning correctly today (a drawback of web-based services).
Some drawbacks of iTOL, at least for my purposes, are:
- Lack of tools/options for displaying or editing the tree itself. You can have rectangular or circular dendrograms, but you can’t control the look of the tree (e.g. colouring branches or changing branch weights, rotating subtrees, controlling ladderizing, etc). Some of this can be got around by editing the newick file first in some other program, and changing colours and weights later by adjusting the PDF output by iTOL. But this is fiddly and I find the SVG graphics in the iTOL PDFs hard to manipulate in Illustrator.
- You can’t display multiple heatmaps on the same figure. With colour strips, you can tick the ‘prevent overlap’ box so that the colour strip is presented adjacent to the previous one rather than over the top of it… so if I have one colour strip representing the location of the isolate, and one representing the presence of different drug resistance mutations, these strips can appear together, side-by-side on the same graph. On the other hand, if I have a heatmap showing the presence of a set of resistance genes, and a second heatmap showing say the MICs of certain drugs, I can’t display these next to each other on the same plot, but only one at a time.
- It is web-based. While there are some advantages to this, I find it frustrating to have to upload and download things all the time, especially for a program like this that doesn’t actually need to access any other databases or compute clusters. You can however establish a private account, and keep track of your trees in separate projects and groups, which is nice. There is also an API and batch access, if you want to link it in with other web services.
ETE2 Python module
Another option is the ETE2 module for Python. This is essentially a package for navigating and displaying phylogenetic trees, and it has some really great features. You need to have some familiarity with Python to be able to use this, but if that’s not a problem for you, then it is worth delving into. It allows you to display sequence alignments, images, protein domains, heatmaps, graphs etc overlaid on tree nodes, leaves or next to the tree, and has the option of half-circle tree plots in addition to the usual full circle tree layout. For examples see the web page. ETE comes with its own graphical interface for displaying trees, as well as being able to write them out directly to image files.
ETE2 is incredibly flexible and I can do just about everything I want with it… EXCEPT, to enforce tree ladderizing when plotting a tree alongside a heatmap. It can ladderized trees, but not when you are using the heatmap display. So unless your tree happens to be naturally ladderized, it can look a bit strange. This is very annoying, as for most of my data, the phylogenetic structure is much clearer when the tree is ladderized, like this:
Anyway, assuming ladderizing isn’t of great concern, it is easy to plot a heatmap next to a tree. Like iTOL, you just need a tree in newick format, and a matrix representing your heatmap. Unlike iTOL, you need to make sure first that the names in your tree and matrix match up 1:1. Also, your matrix should be tab-separated and the first column (which contains the leaf names) should be titled ‘#Names’.
The following commands will give then display your tree and heatmap:
from ete2 import Tree, ClusterTree
t2 = ClusterTree(“tree.nwk”, text_array=”matrix.txt”)
t2.show(“heatmap”)
You can also colour in the leaf nodes and add other data, akin to iTOL, using ETE2. At the end of the post are some example code for how to colour in the leaf nodes with location data, and add a colour strip indicating our resistance mutations, based on the ETE2 tutorial.
Drawbacks of ETE2, for my purposes:
- As with iTOL, there is not adequate control over the layout of the tree, so we can’t get nicely ladderized trees. I also had problems with the branch leading from the root of the tree being drawn as super long by ETE, which makes it awkward to render as a nice image.
- For heatmaps, you have virtually no control over the colour scheme. There are 3 options, from the ETE2 reference “0=green & blue; 1=green & red; 2=red & blue. In all three cases, missing values are rendered in black and transition color (values=center) is white.” However when I tried colourscheme 2 (red & blue) with binary data (0, 1), I got 0 showing up as black and 1 showing up as red, however I specified the minimum, maximum and center values.
- For me, using Python is not a drawback but mostly a strength, as it allows fine control over some of the aspects of the display, and it means you aren’t relying on web services functioning well, or uploading and downloading files. It can also facilitate building the tree drawing into other data analysis pipelines (much like iTOL batch could be, I suppose). But for many people this will be a hindrance.
R packages/functions
(For those of you don’t know R, what follows will not make much sense… maybe look at the pictures to decide if it’s worth learning more about R!)
Heatmaps can be displayed easily in R using the ‘image’ function, and there are loads of heatmap functions that will take a data matrix, cluster the rows and/or columns of the matrix and display the reordered heatmap (plotted using the ‘image’ function on the reordered matrix) alongside a dendrogram representing the clustering. However, I haven’t found a version of this function that allows you to pre-specify the dendrogram as a phylo object, which would allow us to easily display a predefined phylogenetic tree alongside a heatmap representation of associated data.
The R class ‘phylo4d’ does kind of what I want. It can contain a phylogenetic tree and data matrix, mainly for analysis rather than data display, although there are a couple of functions for plotting the data against the tree. The most versatile I could find is ‘table.phylo4d’ function in the ‘adephylo’ package. However, there is still very little control over the way the data is displayed and it can’t give you an actual heatmap. Instead it represents values in the data matrix by the size of circles or squares laid out in a grid. For example, here is what you get by following the examples in the adephylo manual:
This is OK, but I’d rather have a heatmap than scaled circles.
Although we can’t use the ‘image’ command to get this with adephylo, we can approximate it using ‘symbol=”colors”‘ in the table.phylo4d function, setting the plotted points to squares using ‘pch=15’ and specifying a colour set using ‘cols=X”. Here it is with my data, using a simple black=present, white=absent colour scheme:
library(adephylo)
m<-read.delim(“matrix.txt”,row.names=1)
t<-ladderize(read.tree(“tree.nwk”))
p<-phylo4d(t,m)
table.phylo4d(p,symbol=”colors”,scale=F,col=c(“white”,”black”),legend=F,grid=F,show.tip.label=F,ratio.tree=0.25,var.label=1:36,cex.symbol=1,pch=15,box=F)
Yay this gives me proper ladderizing!
Note that you could do much the same with continuous data, just leave the default ‘cols=heat.colors()’ or set it to another range of colours of your choosing, as you might with the regular ‘image’ or ‘heatmap’ functions in R.
In this example, I’ve switched off printing of the leaf labels, but you could switch this back on using ‘show.tip.label=T’. Also in this example I’ve specified that the tree should take up 25% of the width of the image, but you can change this by changing ‘ratio.tree=0.25’ to something else.
Note I’ve coloured the leaves of the tree according to location, using a file with the leaf names in column 1 and locations (categories) in column 2:
tips<-t$edge[,2]
tip.order<-tips[tips<=length(t$tip.label)]
tip.label.order<-t$tip.label[tip.order]
loc<-read.csv(“locations.csv”,row.names=1)
loc1<-as.matrix(loc)[row.names(loc) %in% t$tip.label,] #vector
tipLabelSet <- character(length(loc1))
names(tipLabelSet) <- names(loc1)
groups<-table(loc1)
n<-length(groups)
groupNames<-names(groups)
colours<-rainbow(n) # or set to colours of your choice
for (i in 1:n) {
g<-groupNames[i]
tipLabelSet[loc1==g]<-colours[i]
}
table.phylo4d(p,symbol=”colors”,scale=F,col=c(“white”,”black”),legend=F,grid=F,show.tip.label=F,ratio.tree=0.25,var.label=1:36,cex.symbol=1,pch=15,box=F)tiplabels(col= tipLabelSet[tl$tip.label],pch=16,cex=2)
Note that you can create PDFs in R by preceding the plotting commands with a call to the pdf function and following it with a call to close the graphics device , like this:
pdf(file=”tree.pdf”,10,20) # width=10 inches, height=20 inches
table.phylo4d(p,symbol=”colors”,scale=F,col=c(“white”,”black”),legend=F,grid=F,show.tip.label=F,ratio.tree=0.25,var.label=1:36,cex.symbol=1,pch=15,box=F)
tiplabels(col= tipLabelSet[tl$tip.label],pch=16,cex=2)
dev.off()
Drawbacks of phylo4d plotting in R:
- This is not really a heatmap, and unlike the heatmap or image functions you will need to play with the layout – including symbol size (cex.symbol) and the size of the drawing device in order to get something that looks good and renders all the data points visible.
My simple R function for plotting a heatmap against a tree
While the phylo4d option is pretty good, I figure that since I’m in R, I should be able to use the more powerful ‘image’ function to draw a proper heatmap alongside my tree. So I wrote a little R function to achieve this, called plotTreeData. It uses the ‘ape’ library to read and plot the tree, so you’ll need to have this installed. To use the function, just download the text file here and load it into R like this:
source(“plotTreeData.R”)
The required inputs are:
- treeFile (path to tree file in newick format, tip labels must match those in the data file)
- matrixFile (path to matrix file in csv format, column 1 must contain strain identifiers that match the tip labels in the tree file; other columns contain the data for plotting as a heatmap; column names should be provided. Note that if you already have the matrix loaded into R (e.g. if you have created or manipulated it in R) you can just provide the R object here.)
e.g. plotTreeData(“tree.nwk”,”matrix.csv”) will generate a figure like this, with a greyscale heatmap, in the R graphics device:
To write the image to a PDF or PNG file, provide a file name for the figure via ‘outputPDF=’ or ‘outputPNG=’ (note you can’t do both at once). You can optionally provide width and height using ‘w=’ and ‘h=’ otherwise the defaults for pdf() or png() functions will be used (note for PDF, the units are in inches while for PNG, the units are in pixels).
So to render a figure as a PDF: plotTreeData(“tree.nwk”,”matrix.csv”,outputPDF=”tree.pdf”,w=10,h=20)
Other options include:
- matrix.colours: By default, the heatmap is rendered in greyscale (white to black), but this can be changed using ‘matrix.colours=’, e.g. matrix.colours=heat.colors(10).
function(treeFile,matrixFile,infoFile=NULL,locFile=NULL,outputPDF=NULL,outputPNG=NULL,w,h,matrix.colours=rev(gray(seq(0,1,0.1))),matrix.legend=F,tip.labels=F,offset=0,tip.colour.cex=0.5,legend=T,legend.pos=”bottomleft”,ancestral.reconstruction=F,boundaries=c(0.5,0.75),cluster=F,locColours=NULL,lwd=1.5)
————
Colouring nodes and adding data in ETE2:
We can also colour in the leaves of the tree according to geographic location, using the same input file as for iTOL colour strip:
from ete2 import NodeStyle, TreeStyle
t= Tree(“tree.nwk”)
location_colours = {}
f = file(“locations.csv”)
for line in f:
fields=line.rstrip().split(“,”)
location_colours[fields[0]] = fields[1]# Basic tree style
ts = TreeStyle()
ts.show_leaf_name = True# Create an independent node style for each leaf, which
# specifies the colour given in the locations.csv file
for n in t.get_leaves():
name = n.get_leaf_names()[0]
nstyle = NodeStyle()
nstyle[“fgcolor”] = location_colours[name]
nstyle[“size”] = 10
n.set_style(nstyle)# Let’s now modify the aspect of the root node
t.img_style[“size”] = 30
t.img_style[“fgcolor”] = “blue”t.show(tree_style=ts)
To get both the heatmap and the node colouring, we need to create a new ProfileFace (rather than using the default “heatmap” one) that includes both the heatmap and a coloured circle at each leaf indicating the location (see figure below):
import numpy
from ete2 import ClusterTree, TreeStyle, AttrFace, ProfileFace, TextFace
from ete2.treeview.faces import add_face_to_nodet = ClusterTree(“tree.nwk”, text_array=”matrix.txt”)
array = t.arraytable
# Calculates some stats on the matrix. Needed to establish the heatmap color
# gradients.
matrix_dist = [i for r in xrange(len(array.matrix))\
for i in array.matrix[r] if numpy.isfinite(i)]
matrix_max = numpy.max(matrix_dist)
matrix_min = numpy.min(matrix_dist)
matrix_avg = matrix_min+((matrix_max-matrix_min)/2)# Creates a profile face that will represent node’s profile as a
# heatmap
profileFace = ProfileFace(matrix_max, matrix_min, matrix_avg, \
200, 14, “heatmap”)# Creates your own layout function that uses previous faces
def mylayout(node):
# If node is a leaf
if node.is_leaf():
# Add heatmap line
add_face_to_node(profileFace, node, 0, aligned=True)# Colour in node according to location
node.img_style[“size”]=10
name = node.get_leaf_names()[0]
node.img_style[“fgcolor”] = location_colours[name]ts = TreeStyle()
ts.layout_fn = mylayout
t.show(tree_style=ts)
Now, we have locations shown at the leaves; a heatmap displaying properly; what about showing other variables, like resistance mutations, similar to a colour strip in iTOL?
This can be done as an additional CircleFace, next to the ProfileFace that is holding the heatmap. You can control the order of these using the ‘column’ parameter in the add_face_to_node function:
‘add_face_to_node(face, node, column, aligned=False, position=’branch-right’)‘
So, we just need to add in a few lines to read in another data set:
mutation_colours = {}
f = file(“gyrA_itol_colourstrip.csv”)
for line in f:
fields=line.rstrip().split(“,”)
mutation_colours[fields[0]] = fields[1]
And add an extra ‘add_face_to_node’ line to our layout definition:
def mylayout(node):
# If node is a leaf
if node.is_leaf():
# Add a heatmap line
add_face_to_node(profileFace, node, 0, aligned=True)
# colour node
node.img_style[“size”]=10
name = node.get_leaf_names()[0]
node.img_style[“fgcolor”] = location_colours[name]
# add mutation coloured circle, after the heatmap
add_face_to_node(CircleFace(10, mutation_colours[name], style=’circle’), node, 1, aligned=True)







nice summary & tutorial! Will come in handy.
Thanks for this nice review! Just a few comments about ETE that may be useful:
[1] the bug regarding the long branch leading to the root node is now solved in the development branch. However, as for the current stable release, it can be easily circumvented by setting the root-node distance attribute to zero (t2.dist=0, in your code example).
[2] tree.ladderize() should work also in ClusterTrees (at least it works in my tests). If not, please feel free to open an issue in our support mailing list (http://groups.google.com/group/etetoolkit) and I will be happy to fix it ASAP.
[3] controlling heatmap color scheme is on the roadmap, you might be interested in the following workaround: https://groups.google.com/forum/?fromgroups#!searchin/etetoolkit/color$20scheme/etetoolkit/_3adcV-rBec/TT9dfodkd1EJ
This is really useful
Excellent tutorial…!
What format is the locations.csv file under ‘R packages/functions’ section? Can you please show us an example just to see how it looks like.
Pingback: OpenLab – Libraries to visualize phylogenetic trees
Pingback: Tree viewers and editors | m's Bioinformatics
I found this very helpful – thanks! 🙂
Pingback: Annotating phylogenetic trees with gene presence/absence etc | Bits and Bugs
Hi there, http://www.scriptree.org/ is another tool to visualize annotations in regards of tips of the tree or even at internal labels. Instructions let you specify how you want your newick tree to be displayed as well as how annotations should be displayed